Ubiquitin-Dependent Cell-Fate Decisions During Human Development and Disease
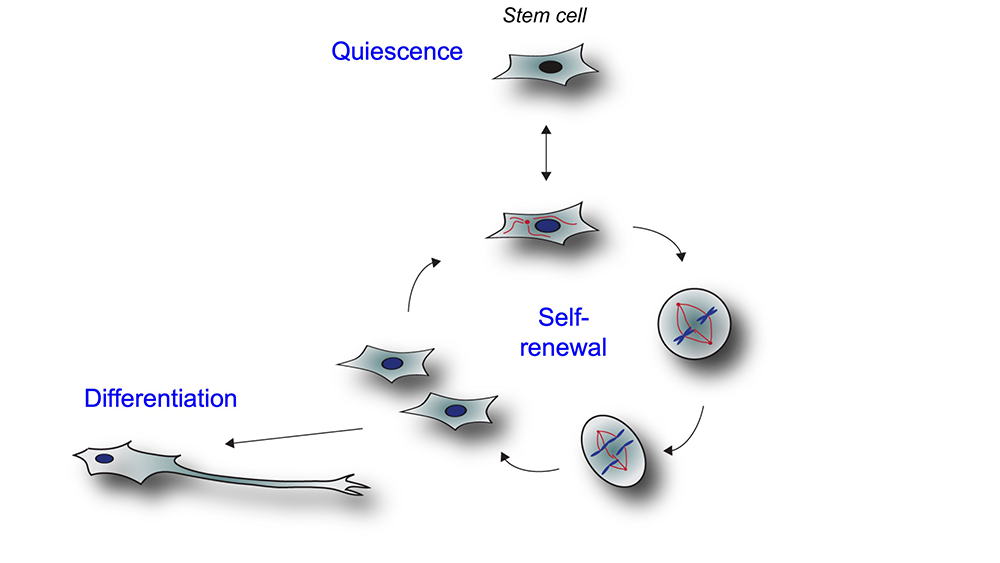
Mammalian development relies on the precise execution of highly coordinated cell-fate decisions by stem cells, which can undergo self-renewal, reversibly exit into a quiescent state, or terminally commit to a cell differentiation program (Figure 1).
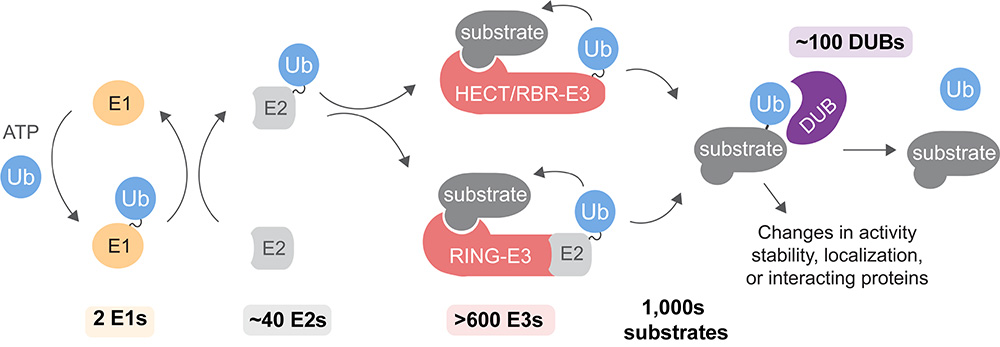
To orchestrate these decisions, stem cells make frequent use of ubiquitylation, an essential post-translational modification that alters the stability, activity, localization, or interaction landscape of target proteins (Figure 2). Ubiquitin is attached to target proteins by ubiquitin E1 activating, E2 conjugating, and E3 ligating enzymes and cleaved off by deubiquitylases (DUBs). We study the molecular mechanisms of how these ubiquitylation enzymes determine cell-fate decisions during differentiation. We focus on those enzymes linked to congenital diseases of impaired brain and craniofacial development or autoinflammation.
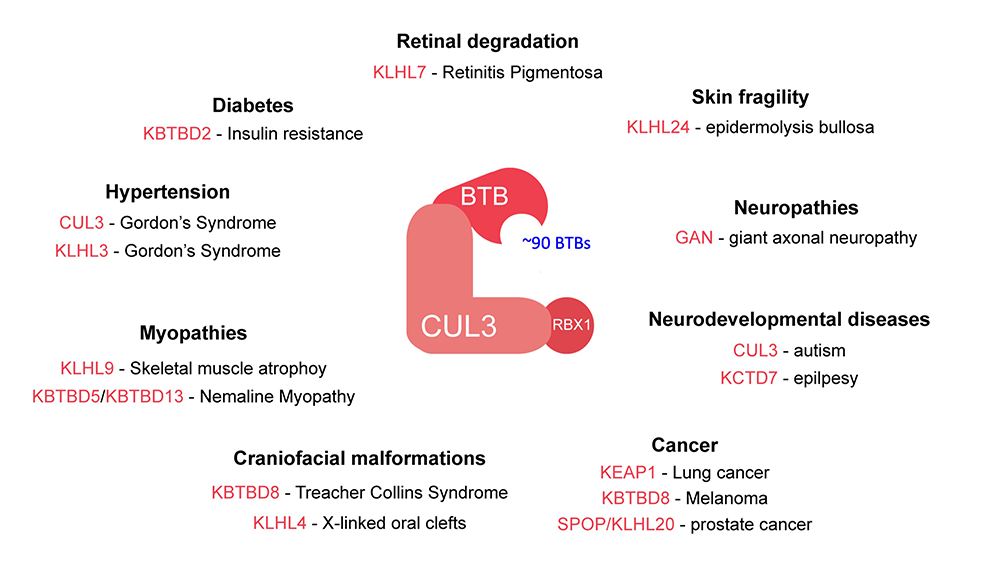
For our studies we leverage the unique clinical environment of the NIH intramural program and its large research hospital, and closely collaborate with human geneticists. This way, we combine our lab’s core expertise in biochemistry, proteomics, and human pluripotent stem cell culture with human genetics to determine the molecular mechanism of how ubiquitylation controls embryonic cell-fate choices. Current projects in our lab center around CUL3-based E3 ligases, a large family of multi-subunit E3s that uses one of ~90 BTB proteins as substrate adaptors. Dysregulation of or mutations in many of these CUL3-BTB complexes lead to human diseases (Figure 3, Asmar, et al., ECR, 2020) and we are eager to elucidate the molecular underpinnings of how this occurs. Examples include the E3 ligase complex CUL3-KBTBD8, which we have shown to regulate signaling pathways that promote neural crest specification (Werner, et al., Nature, 2015; Werner, et al., eLife, 2018) and when dysregulated results in the craniofacial development disease Treacher Collins Syndrome. Similarly, we recently found CUL3-KLHL4 to regulate cytoskeletal signaling networks to control early steps in the development of the brain, face, and skin (Asmar, et al., Nat Commun, 2023). In addition, we are interested in understanding how the DUB OTUD5 regulates chromatin dynamics during early stages of human development and if misregulated leads to a multiple congenital anomaly disease we have named LINKED (LINKage-specific-deubiquitylation-deficiency-induced Embryonic Defects) syndrome (Beck, et al., Sci Adv 2021). Finally, we aim to determine how the ubiquitin activating enzyme UBA1 and regulated ubiquitin activation controls hematopoietic cell-fate decisions and if misregulated leads to an autoinflammatory disease we have termed VEXAS (Vacuoles, E1 enzyme, X-linked, Autoinflammatory, Somatic) syndrome (Beck, et al., 2020, N Engl J Med;Ferrada, et al., 2022, Blood).
We hope that the results of our studies will provide mechanistic insights into important aspects of human development and into the molecular origin of human diseases, which will be useful for developing novel therapeutic approaches.